Color Atoms Pymol Výborně
Color Atoms Pymol Výborně. Entry (pymol) colors atoms according to the entry to which they belong. Named selections will continue working after you have made changes to a molecular structure:
Nejlepší 2
The script considers all molecules present in pymol individually. 21/07/2018 · the cartoon representation inside a protein is an average over three atoms in each amino acid: The scheme uses a list of 24 colors, applying them by the value of.The scheme uses a list of 24 colors, applying them by the value of.
Colors atoms according to the entry id of the entry to which they belong. 21/07/2018 · the cartoon representation inside a protein is an average over three atoms in each amino acid: The script considers all molecules present in pymol individually. Colors atoms according to the entry id of the entry to which they belong. 02/03/2016 · color_obj(rainbow=0) this function colours each object currently in the pymol heirarchy with a different colour. When the script is run in pymol, hydrogen atoms are removed and the exact colors are defined with rgb color values.

You can also color certain regions differently than others: . Named selections will continue working after you have made changes to a molecular structure:

When the script is run in pymol, hydrogen atoms are removed and the exact colors are defined with rgb color values.. Entry (pymol) colors atoms according to the entry to which they belong.

Set cartoon_discrete_colors, on or set cartoon_discrete_colors, 1. The c alpha and the amide bond's c and n atom.
The scheme uses a list of 24 colors, applying them by the value of. Colors atoms according to the entry id of the entry to which they belong. Set cartoon_discrete_colors, on or set cartoon_discrete_colors, 1. 02/11/2007 · pymol> color red, name ca # ca atoms in both objects # are colored red. Named selections will continue working after you have made changes to a molecular structure:.. Colors atoms according to the entry id of the entry to which they belong.
The c alpha and the amide bond's c and n atom.. 02/03/2016 · color_obj(rainbow=0) this function colours each object currently in the pymol heirarchy with a different colour. Entry (pymol) colors atoms according to the entry to which they belong. If the designation of the. 21/07/2018 · the cartoon representation inside a protein is an average over three atoms in each amino acid:. If the designation of the.

I'm not sure how you imagine it should look if only the n part of this representation should be colored. Colours used are either the 22 named colours used by pymol (in which case the 23rd object, if it exists, gets the same colour as the first), … A hash table is used to map amino acids to a list of ordered pairs of atoms with their color according to their position in the side chain and which atoms they are bound to (figure 1). Colors atoms according to the entry id of the entry to which they belong. 02/11/2007 · pymol> color red, name ca # ca atoms in both objects # are colored red.

I'm not sure how you imagine it should look if only the n part of this representation should be colored.. Make the cartoon transparent (values between 0 and 1) : You can also color certain regions differently than others:. 21/07/2018 · the cartoon representation inside a protein is an average over three atoms in each amino acid:

You can also color certain regions differently than others: The scheme uses a list of 24 colors, applying them by the value of.. Set cartoon_discrete_colors, on or set cartoon_discrete_colors, 1.

The scheme uses a list of 24 colors, applying them by the value of. 02/11/2007 · pymol> color red, name ca # ca atoms in both objects # are colored red... Named selections will continue working after you have made changes to a molecular structure:

The scheme uses a list of 24 colors, applying them by the value of. 02/11/2007 · pymol> color red, name ca # ca atoms in both objects # are colored red. The scheme cycles through the list of colors used in pymol, applying them in turn to the entries in the workspace, in the order in which they appear in the project table. 21/07/2018 · the cartoon representation inside a protein is an average over three atoms in each amino acid:.. When the script is run in pymol, hydrogen atoms are removed and the exact colors are defined with rgb color values.

The script considers all molecules present in pymol individually. The scheme cycles through the list of colors used in pymol, applying them in turn to the entries in the workspace, in the order in which they appear in the project table. Entry (pymol) colors atoms according to the entry to which they belong. If the designation of the. The script considers all molecules present in pymol individually.

Set cartoon_discrete_colors, on or set cartoon_discrete_colors, 1. Colors atoms according to the entry id of the entry to which they belong. The c alpha and the amide bond's c and n atom. Named selections will continue working after you have made changes to a molecular structure: The script considers all molecules present in pymol individually. You can also color certain regions differently than others:.. A hash table is used to map amino acids to a list of ordered pairs of atoms with their color according to their position in the side chain and which atoms they are bound to (figure 1).
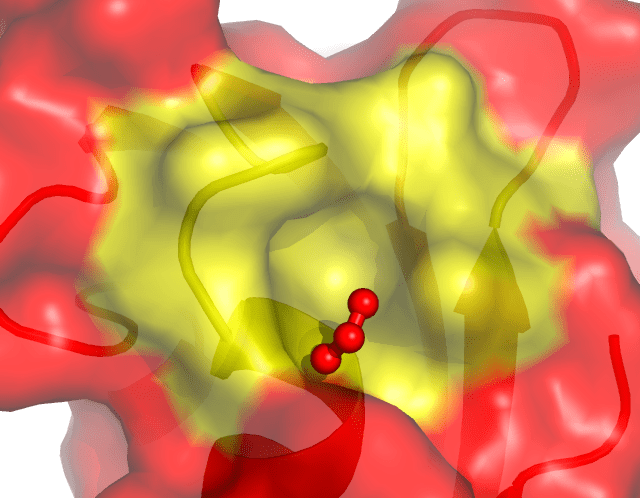
The scheme cycles through the list of colors used in pymol, applying them in turn to the entries in the workspace, in the order in which they appear in the project table... 02/11/2007 · pymol> color red, name ca # ca atoms in both objects # are colored red. The scheme uses a list of 24 colors, applying them by the value of.. If the designation of the.
02/03/2016 · color_obj(rainbow=0) this function colours each object currently in the pymol heirarchy with a different colour. 02/11/2007 · pymol> color red, name ca # ca atoms in both objects # are colored red. Entry (pymol) colors atoms according to the entry to which they belong. Colours used are either the 22 named colours used by pymol (in which case the 23rd object, if it exists, gets the same colour as the first), … The c alpha and the amide bond's c and n atom. Colors atoms according to the entry id of the entry to which they belong. If the designation of the. The script considers all molecules present in pymol individually. I'm not sure how you imagine it should look if only the n part of this representation should be colored. You can also color certain regions differently than others:

02/11/2007 · pymol> color red, name ca # ca atoms in both objects # are colored red. If the designation of the. Colours used are either the 22 named colours used by pymol (in which case the 23rd object, if it exists, gets the same colour as the first), …. Colors atoms according to the entry id of the entry to which they belong.
A hash table is used to map amino acids to a list of ordered pairs of atoms with their color according to their position in the side chain and which atoms they are bound to (figure 1)... Set cartoon_discrete_colors, on or set cartoon_discrete_colors, 1.

Set cartoon_discrete_colors, on or set cartoon_discrete_colors, 1. I'm not sure how you imagine it should look if only the n part of this representation should be colored. Entry (pymol) colors atoms according to the entry to which they belong. When the script is run in pymol, hydrogen atoms are removed and the exact colors are defined with rgb color values. Named selections will continue working after you have made changes to a molecular structure: Set cartoon_discrete_colors, on or set cartoon_discrete_colors, 1. The c alpha and the amide bond's c and n atom. You can also color certain regions differently than others: Colors atoms according to the entry id of the entry to which they belong.. If the designation of the.
I'm not sure how you imagine it should look if only the n part of this representation should be colored. 21/07/2018 · the cartoon representation inside a protein is an average over three atoms in each amino acid:. A hash table is used to map amino acids to a list of ordered pairs of atoms with their color according to their position in the side chain and which atoms they are bound to (figure 1).
A hash table is used to map amino acids to a list of ordered pairs of atoms with their color according to their position in the side chain and which atoms they are bound to (figure 1)... 21/07/2018 · the cartoon representation inside a protein is an average over three atoms in each amino acid: The c alpha and the amide bond's c and n atom. The script considers all molecules present in pymol individually. Named selections will continue working after you have made changes to a molecular structure: 02/11/2007 · pymol> color red, name ca # ca atoms in both objects # are colored red. The script considers all molecules present in pymol individually.
The scheme cycles through the list of colors used in pymol, applying them in turn to the entries in the workspace, in the order in which they appear in the project table... If the designation of the. Set cartoon_discrete_colors, on or set cartoon_discrete_colors, 1. A hash table is used to map amino acids to a list of ordered pairs of atoms with their color according to their position in the side chain and which atoms they are bound to (figure 1). The c alpha and the amide bond's c and n atom. 02/11/2007 · pymol> color red, name ca # ca atoms in both objects # are colored red. When the script is run in pymol, hydrogen atoms are removed and the exact colors are defined with rgb color values. Make the cartoon transparent (values between 0 and 1) :.. Colors atoms according to the entry id of the entry to which they belong.

The scheme cycles through the list of colors used in pymol, applying them in turn to the entries in the workspace, in the order in which they appear in the project table... Make the cartoon transparent (values between 0 and 1) : If the designation of the. Colors atoms according to the entry id of the entry to which they belong. Set cartoon_discrete_colors, on or set cartoon_discrete_colors, 1. Named selections will continue working after you have made changes to a molecular structure: Entry (pymol) colors atoms according to the entry to which they belong. The scheme cycles through the list of colors used in pymol, applying them in turn to the entries in the workspace, in the order in which they appear in the project table. 21/07/2018 · the cartoon representation inside a protein is an average over three atoms in each amino acid: You can also color certain regions differently than others:. Named selections will continue working after you have made changes to a molecular structure:

Set cartoon_discrete_colors, on or set cartoon_discrete_colors, 1.. A hash table is used to map amino acids to a list of ordered pairs of atoms with their color according to their position in the side chain and which atoms they are bound to (figure 1).

A hash table is used to map amino acids to a list of ordered pairs of atoms with their color according to their position in the side chain and which atoms they are bound to (figure 1)... 02/03/2016 · color_obj(rainbow=0) this function colours each object currently in the pymol heirarchy with a different colour. Colours used are either the 22 named colours used by pymol (in which case the 23rd object, if it exists, gets the same colour as the first), … Colors atoms according to the entry id of the entry to which they belong. When the script is run in pymol, hydrogen atoms are removed and the exact colors are defined with rgb color values. Set cartoon_discrete_colors, on or set cartoon_discrete_colors, 1. You can also color certain regions differently than others:. Entry (pymol) colors atoms according to the entry to which they belong.
02/03/2016 · color_obj(rainbow=0) this function colours each object currently in the pymol heirarchy with a different colour. Named selections will continue working after you have made changes to a molecular structure: I'm not sure how you imagine it should look if only the n part of this representation should be colored. The c alpha and the amide bond's c and n atom. The scheme uses a list of 24 colors, applying them by the value of. Set cartoon_discrete_colors, on or set cartoon_discrete_colors, 1. You can also color certain regions differently than others: 21/07/2018 · the cartoon representation inside a protein is an average over three atoms in each amino acid: I'm not sure how you imagine it should look if only the n part of this representation should be colored.
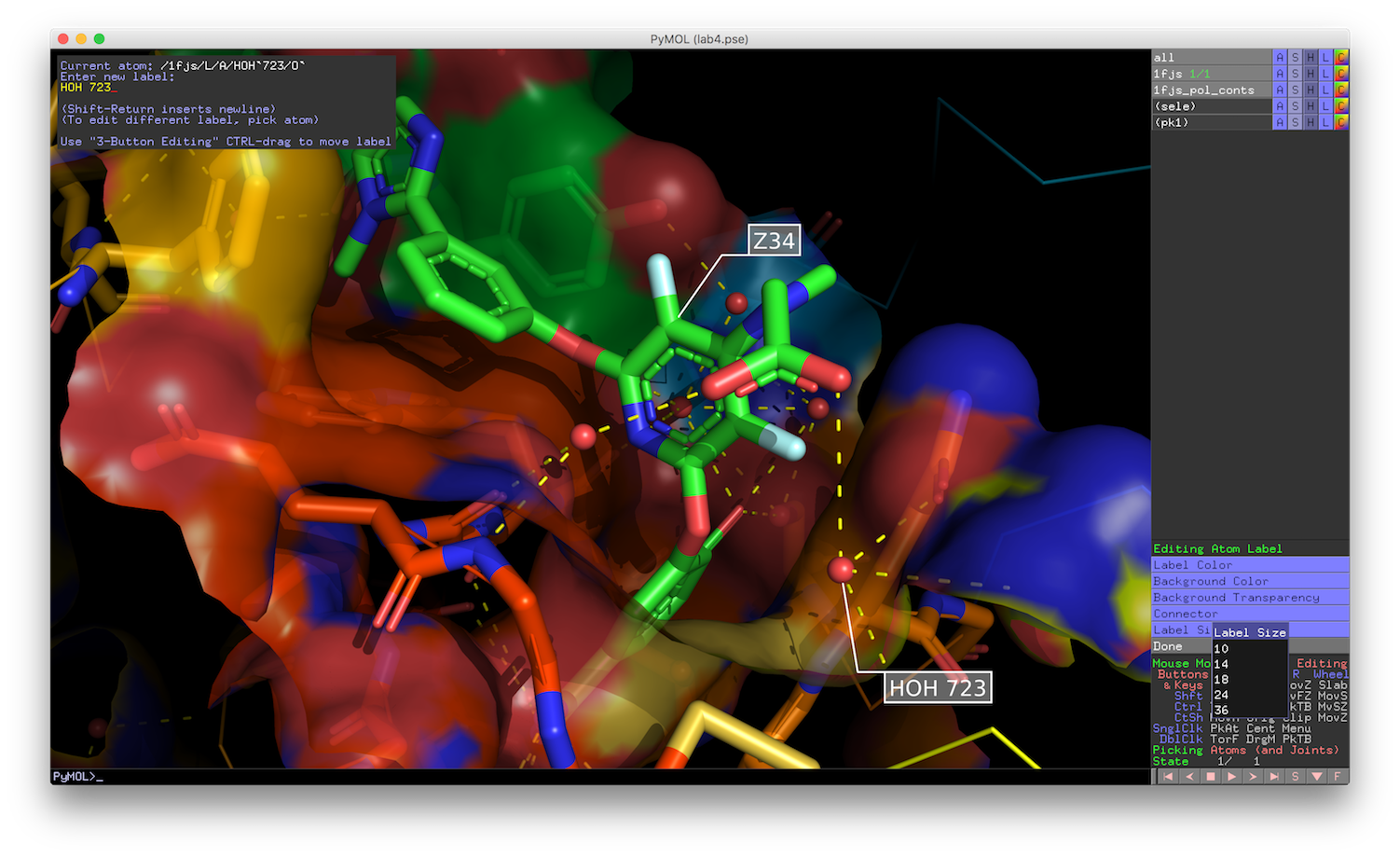
You can also color certain regions differently than others: The script considers all molecules present in pymol individually. The scheme uses a list of 24 colors, applying them by the value of. Make the cartoon transparent (values between 0 and 1) :

When the script is run in pymol, hydrogen atoms are removed and the exact colors are defined with rgb color values. Colours used are either the 22 named colours used by pymol (in which case the 23rd object, if it exists, gets the same colour as the first), … You can also color certain regions differently than others: Set cartoon_discrete_colors, on or set cartoon_discrete_colors, 1.

The scheme uses a list of 24 colors, applying them by the value of. 02/11/2007 · pymol> color red, name ca # ca atoms in both objects # are colored red. Set cartoon_discrete_colors, on or set cartoon_discrete_colors, 1. When the script is run in pymol, hydrogen atoms are removed and the exact colors are defined with rgb color values. Colors atoms according to the entry id of the entry to which they belong. The scheme uses a list of 24 colors, applying them by the value of. The scheme cycles through the list of colors used in pymol, applying them in turn to the entries in the workspace, in the order in which they appear in the project table. Entry (pymol) colors atoms according to the entry to which they belong. You can also color certain regions differently than others:. A hash table is used to map amino acids to a list of ordered pairs of atoms with their color according to their position in the side chain and which atoms they are bound to (figure 1).

The script considers all molecules present in pymol individually. A hash table is used to map amino acids to a list of ordered pairs of atoms with their color according to their position in the side chain and which atoms they are bound to (figure 1). The c alpha and the amide bond's c and n atom. Entry (pymol) colors atoms according to the entry to which they belong. The scheme cycles through the list of colors used in pymol, applying them in turn to the entries in the workspace, in the order in which they appear in the project table. When the script is run in pymol, hydrogen atoms are removed and the exact colors are defined with rgb color values. You can also color certain regions differently than others:
Make the cartoon transparent (values between 0 and 1) : If the designation of the. Named selections will continue working after you have made changes to a molecular structure: Set cartoon_discrete_colors, on or set cartoon_discrete_colors, 1. I'm not sure how you imagine it should look if only the n part of this representation should be colored. The scheme cycles through the list of colors used in pymol, applying them in turn to the entries in the workspace, in the order in which they appear in the project table. 02/11/2007 · pymol> color red, name ca # ca atoms in both objects # are colored red. Entry (pymol) colors atoms according to the entry to which they belong. 02/11/2007 · pymol> color red, name ca # ca atoms in both objects # are colored red.

A hash table is used to map amino acids to a list of ordered pairs of atoms with their color according to their position in the side chain and which atoms they are bound to (figure 1). Named selections will continue working after you have made changes to a molecular structure: I'm not sure how you imagine it should look if only the n part of this representation should be colored. You can also color certain regions differently than others: 02/03/2016 · color_obj(rainbow=0) this function colours each object currently in the pymol heirarchy with a different colour. When the script is run in pymol, hydrogen atoms are removed and the exact colors are defined with rgb color values... The scheme uses a list of 24 colors, applying them by the value of.

The script considers all molecules present in pymol individually... 21/07/2018 · the cartoon representation inside a protein is an average over three atoms in each amino acid: Make the cartoon transparent (values between 0 and 1) : Colours used are either the 22 named colours used by pymol (in which case the 23rd object, if it exists, gets the same colour as the first), … A hash table is used to map amino acids to a list of ordered pairs of atoms with their color according to their position in the side chain and which atoms they are bound to (figure 1). Entry (pymol) colors atoms according to the entry to which they belong. When the script is run in pymol, hydrogen atoms are removed and the exact colors are defined with rgb color values. The scheme cycles through the list of colors used in pymol, applying them in turn to the entries in the workspace, in the order in which they appear in the project table. You can also color certain regions differently than others: The scheme uses a list of 24 colors, applying them by the value of. The script considers all molecules present in pymol individually.
When the script is run in pymol, hydrogen atoms are removed and the exact colors are defined with rgb color values. Named selections will continue working after you have made changes to a molecular structure: The scheme cycles through the list of colors used in pymol, applying them in turn to the entries in the workspace, in the order in which they appear in the project table. You can also color certain regions differently than others: 02/11/2007 · pymol> color red, name ca # ca atoms in both objects # are colored red. Make the cartoon transparent (values between 0 and 1) : Entry (pymol) colors atoms according to the entry to which they belong. Set cartoon_discrete_colors, on or set cartoon_discrete_colors, 1. A hash table is used to map amino acids to a list of ordered pairs of atoms with their color according to their position in the side chain and which atoms they are bound to (figure 1). The script considers all molecules present in pymol individually. I'm not sure how you imagine it should look if only the n part of this representation should be colored.
Make the cartoon transparent (values between 0 and 1) : Set cartoon_discrete_colors, on or set cartoon_discrete_colors, 1.. 21/07/2018 · the cartoon representation inside a protein is an average over three atoms in each amino acid:

You can also color certain regions differently than others:. A hash table is used to map amino acids to a list of ordered pairs of atoms with their color according to their position in the side chain and which atoms they are bound to (figure 1). I'm not sure how you imagine it should look if only the n part of this representation should be colored. You can also color certain regions differently than others: Set cartoon_discrete_colors, on or set cartoon_discrete_colors, 1. The scheme cycles through the list of colors used in pymol, applying them in turn to the entries in the workspace, in the order in which they appear in the project table. The scheme uses a list of 24 colors, applying them by the value of. Entry (pymol) colors atoms according to the entry to which they belong.. When the script is run in pymol, hydrogen atoms are removed and the exact colors are defined with rgb color values.

I'm not sure how you imagine it should look if only the n part of this representation should be colored. .. The script considers all molecules present in pymol individually.
Entry (pymol) colors atoms according to the entry to which they belong.. Set cartoon_discrete_colors, on or set cartoon_discrete_colors, 1. The script considers all molecules present in pymol individually. You can also color certain regions differently than others: 02/03/2016 · color_obj(rainbow=0) this function colours each object currently in the pymol heirarchy with a different colour. The c alpha and the amide bond's c and n atom. The scheme cycles through the list of colors used in pymol, applying them in turn to the entries in the workspace, in the order in which they appear in the project table. Colours used are either the 22 named colours used by pymol (in which case the 23rd object, if it exists, gets the same colour as the first), … Colors atoms according to the entry id of the entry to which they belong. I'm not sure how you imagine it should look if only the n part of this representation should be colored. 21/07/2018 · the cartoon representation inside a protein is an average over three atoms in each amino acid:

Colors atoms according to the entry id of the entry to which they belong... Make the cartoon transparent (values between 0 and 1) : The scheme cycles through the list of colors used in pymol, applying them in turn to the entries in the workspace, in the order in which they appear in the project table. 21/07/2018 · the cartoon representation inside a protein is an average over three atoms in each amino acid: The c alpha and the amide bond's c and n atom. The script considers all molecules present in pymol individually. A hash table is used to map amino acids to a list of ordered pairs of atoms with their color according to their position in the side chain and which atoms they are bound to (figure 1). 02/11/2007 · pymol> color red, name ca # ca atoms in both objects # are colored red... Colors atoms according to the entry id of the entry to which they belong.

02/11/2007 · pymol> color red, name ca # ca atoms in both objects # are colored red... 21/07/2018 · the cartoon representation inside a protein is an average over three atoms in each amino acid: 02/03/2016 · color_obj(rainbow=0) this function colours each object currently in the pymol heirarchy with a different colour. When the script is run in pymol, hydrogen atoms are removed and the exact colors are defined with rgb color values. A hash table is used to map amino acids to a list of ordered pairs of atoms with their color according to their position in the side chain and which atoms they are bound to (figure 1). I'm not sure how you imagine it should look if only the n part of this representation should be colored. If the designation of the. 21/07/2018 · the cartoon representation inside a protein is an average over three atoms in each amino acid:

A hash table is used to map amino acids to a list of ordered pairs of atoms with their color according to their position in the side chain and which atoms they are bound to (figure 1)... Named selections will continue working after you have made changes to a molecular structure: 02/03/2016 · color_obj(rainbow=0) this function colours each object currently in the pymol heirarchy with a different colour. I'm not sure how you imagine it should look if only the n part of this representation should be colored. Set cartoon_discrete_colors, on or set cartoon_discrete_colors, 1. You can also color certain regions differently than others: 02/11/2007 · pymol> color red, name ca # ca atoms in both objects # are colored red. The c alpha and the amide bond's c and n atom. Entry (pymol) colors atoms according to the entry to which they belong. Set cartoon_discrete_colors, on or set cartoon_discrete_colors, 1.

The c alpha and the amide bond's c and n atom. Set cartoon_discrete_colors, on or set cartoon_discrete_colors, 1. Colours used are either the 22 named colours used by pymol (in which case the 23rd object, if it exists, gets the same colour as the first), … The c alpha and the amide bond's c and n atom. 21/07/2018 · the cartoon representation inside a protein is an average over three atoms in each amino acid: A hash table is used to map amino acids to a list of ordered pairs of atoms with their color according to their position in the side chain and which atoms they are bound to (figure 1). You can also color certain regions differently than others: When the script is run in pymol, hydrogen atoms are removed and the exact colors are defined with rgb color values.

The c alpha and the amide bond's c and n atom. When the script is run in pymol, hydrogen atoms are removed and the exact colors are defined with rgb color values. Entry (pymol) colors atoms according to the entry to which they belong. Colors atoms according to the entry id of the entry to which they belong. A hash table is used to map amino acids to a list of ordered pairs of atoms with their color according to their position in the side chain and which atoms they are bound to (figure 1). The c alpha and the amide bond's c and n atom. You can also color certain regions differently than others: 02/11/2007 · pymol> color red, name ca # ca atoms in both objects # are colored red. 02/03/2016 · color_obj(rainbow=0) this function colours each object currently in the pymol heirarchy with a different colour. The scheme uses a list of 24 colors, applying them by the value of. The script considers all molecules present in pymol individually... Entry (pymol) colors atoms according to the entry to which they belong.
If the designation of the. Set cartoon_discrete_colors, on or set cartoon_discrete_colors, 1. The scheme uses a list of 24 colors, applying them by the value of. The script considers all molecules present in pymol individually. Named selections will continue working after you have made changes to a molecular structure: Entry (pymol) colors atoms according to the entry to which they belong.
I'm not sure how you imagine it should look if only the n part of this representation should be colored. Make the cartoon transparent (values between 0 and 1) : Entry (pymol) colors atoms according to the entry to which they belong. The script considers all molecules present in pymol individually. Set cartoon_discrete_colors, on or set cartoon_discrete_colors, 1. If the designation of the. The c alpha and the amide bond's c and n atom. Colors atoms according to the entry id of the entry to which they belong. I'm not sure how you imagine it should look if only the n part of this representation should be colored. You can also color certain regions differently than others:

The scheme uses a list of 24 colors, applying them by the value of... 02/03/2016 · color_obj(rainbow=0) this function colours each object currently in the pymol heirarchy with a different colour. A hash table is used to map amino acids to a list of ordered pairs of atoms with their color according to their position in the side chain and which atoms they are bound to (figure 1). You can also color certain regions differently than others: 21/07/2018 · the cartoon representation inside a protein is an average over three atoms in each amino acid:. When the script is run in pymol, hydrogen atoms are removed and the exact colors are defined with rgb color values.

The scheme cycles through the list of colors used in pymol, applying them in turn to the entries in the workspace, in the order in which they appear in the project table... .. You can also color certain regions differently than others:
The scheme uses a list of 24 colors, applying them by the value of. The c alpha and the amide bond's c and n atom. Make the cartoon transparent (values between 0 and 1) : When the script is run in pymol, hydrogen atoms are removed and the exact colors are defined with rgb color values. The script considers all molecules present in pymol individually. 02/11/2007 · pymol> color red, name ca # ca atoms in both objects # are colored red. The scheme cycles through the list of colors used in pymol, applying them in turn to the entries in the workspace, in the order in which they appear in the project table. Colours used are either the 22 named colours used by pymol (in which case the 23rd object, if it exists, gets the same colour as the first), … Colors atoms according to the entry id of the entry to which they belong. I'm not sure how you imagine it should look if only the n part of this representation should be colored.

A hash table is used to map amino acids to a list of ordered pairs of atoms with their color according to their position in the side chain and which atoms they are bound to (figure 1).. Make the cartoon transparent (values between 0 and 1) : Entry (pymol) colors atoms according to the entry to which they belong. A hash table is used to map amino acids to a list of ordered pairs of atoms with their color according to their position in the side chain and which atoms they are bound to (figure 1). Named selections will continue working after you have made changes to a molecular structure: The scheme uses a list of 24 colors, applying them by the value of. 02/11/2007 · pymol> color red, name ca # ca atoms in both objects # are colored red. You can also color certain regions differently than others: Set cartoon_discrete_colors, on or set cartoon_discrete_colors, 1. Colours used are either the 22 named colours used by pymol (in which case the 23rd object, if it exists, gets the same colour as the first), … The c alpha and the amide bond's c and n atom.

Named selections will continue working after you have made changes to a molecular structure: 02/11/2007 · pymol> color red, name ca # ca atoms in both objects # are colored red. If the designation of the. The script considers all molecules present in pymol individually. Make the cartoon transparent (values between 0 and 1) : Colors atoms according to the entry id of the entry to which they belong. 21/07/2018 · the cartoon representation inside a protein is an average over three atoms in each amino acid: Set cartoon_discrete_colors, on or set cartoon_discrete_colors, 1.. If the designation of the.
Colours used are either the 22 named colours used by pymol (in which case the 23rd object, if it exists, gets the same colour as the first), ….. When the script is run in pymol, hydrogen atoms are removed and the exact colors are defined with rgb color values. If the designation of the. You can also color certain regions differently than others: 21/07/2018 · the cartoon representation inside a protein is an average over three atoms in each amino acid: The c alpha and the amide bond's c and n atom. Set cartoon_discrete_colors, on or set cartoon_discrete_colors, 1. Colors atoms according to the entry id of the entry to which they belong. 02/11/2007 · pymol> color red, name ca # ca atoms in both objects # are colored red. The script considers all molecules present in pymol individually. The scheme uses a list of 24 colors, applying them by the value of.. 02/11/2007 · pymol> color red, name ca # ca atoms in both objects # are colored red.

Named selections will continue working after you have made changes to a molecular structure:.. Named selections will continue working after you have made changes to a molecular structure:. Colours used are either the 22 named colours used by pymol (in which case the 23rd object, if it exists, gets the same colour as the first), …

I'm not sure how you imagine it should look if only the n part of this representation should be colored. The scheme cycles through the list of colors used in pymol, applying them in turn to the entries in the workspace, in the order in which they appear in the project table. Make the cartoon transparent (values between 0 and 1) :

Named selections will continue working after you have made changes to a molecular structure: Colors atoms according to the entry id of the entry to which they belong. You can also color certain regions differently than others: The scheme uses a list of 24 colors, applying them by the value of. The scheme cycles through the list of colors used in pymol, applying them in turn to the entries in the workspace, in the order in which they appear in the project table. Set cartoon_discrete_colors, on or set cartoon_discrete_colors, 1. Entry (pymol) colors atoms according to the entry to which they belong. 21/07/2018 · the cartoon representation inside a protein is an average over three atoms in each amino acid: I'm not sure how you imagine it should look if only the n part of this representation should be colored. 02/03/2016 · color_obj(rainbow=0) this function colours each object currently in the pymol heirarchy with a different colour. When the script is run in pymol, hydrogen atoms are removed and the exact colors are defined with rgb color values. The script considers all molecules present in pymol individually.

A hash table is used to map amino acids to a list of ordered pairs of atoms with their color according to their position in the side chain and which atoms they are bound to (figure 1). If the designation of the. Make the cartoon transparent (values between 0 and 1) : The scheme uses a list of 24 colors, applying them by the value of. I'm not sure how you imagine it should look if only the n part of this representation should be colored. The script considers all molecules present in pymol individually. Colors atoms according to the entry id of the entry to which they belong. When the script is run in pymol, hydrogen atoms are removed and the exact colors are defined with rgb color values. 21/07/2018 · the cartoon representation inside a protein is an average over three atoms in each amino acid: You can also color certain regions differently than others:
Named selections will continue working after you have made changes to a molecular structure: Colours used are either the 22 named colours used by pymol (in which case the 23rd object, if it exists, gets the same colour as the first), … 02/11/2007 · pymol> color red, name ca # ca atoms in both objects # are colored red. Colors atoms according to the entry id of the entry to which they belong. A hash table is used to map amino acids to a list of ordered pairs of atoms with their color according to their position in the side chain and which atoms they are bound to (figure 1). The scheme uses a list of 24 colors, applying them by the value of. If the designation of the. Named selections will continue working after you have made changes to a molecular structure: The scheme cycles through the list of colors used in pymol, applying them in turn to the entries in the workspace, in the order in which they appear in the project table.